In the world of rapidly evolving sciences, new discoveries and industry breakthroughs, the emphasis is always on the next ‘big thing’, but what if this next ‘big thing’ was actually smaller than the eye could see? Like DNA? In the last two decades, the cost and efficiency of the molecular ‘toolkit’ for monitoring biodiversity and species distribution has undergone significant improvement, giving rise to environmental DNA (eDNA) metabarcoding [1].
DNA, Deoxyribonucleic Acid, is comprised of two strands of alternating sugars and phosphate groups bound together by base pairs (adenine (A), cytosine (C), guanine (G), and thymine (T)) to form a double helix structure. These base pairs serve as instructions for assembling proteins and are therefore the building blocks for all organisms on the planet [2]. While the chemical structure of DNA is the same for most organisms, differences exist in the order in which the base pairs (G, C, T, A) are presented, and this unique sequence can be used for species identification [3].
As living organisms move through their environment, they deposit genetic material in the form of excretion, skin cells or scales, hair/fur, blood and gametes [4]. This eDNA can be extracted from an environmental sample (water, soil, ice, air), and amplified through polymerase chain reaction (PCR). The amplicons are then sequenced, and these sequences are queried against a database of taxonomically assigned sequences, to determine what species the DNA belongs to [5].

Advantages of eDNA
eDNA metabarcoding provides a rapid, non-invasive, and cost-effective insight into what species are present in the sampled environment, without relying on morphological observations for species identification [5]. These observations, which are associated with traditional monitoring techniques (field signs e.g., hair/feathers, scats, tracks, shelter, feeding signs and carcasses/bones), can be problematic given some species exhibit different morphologies at different life stages and between different environments [6, 1]. As a result, traditional monitoring techniques may lack the resolution to detect high priority species such as rare, endangered, elusive or invasive species, and can be labour intensive and expensive when applied over large spatial and temporal scales [7, 6].
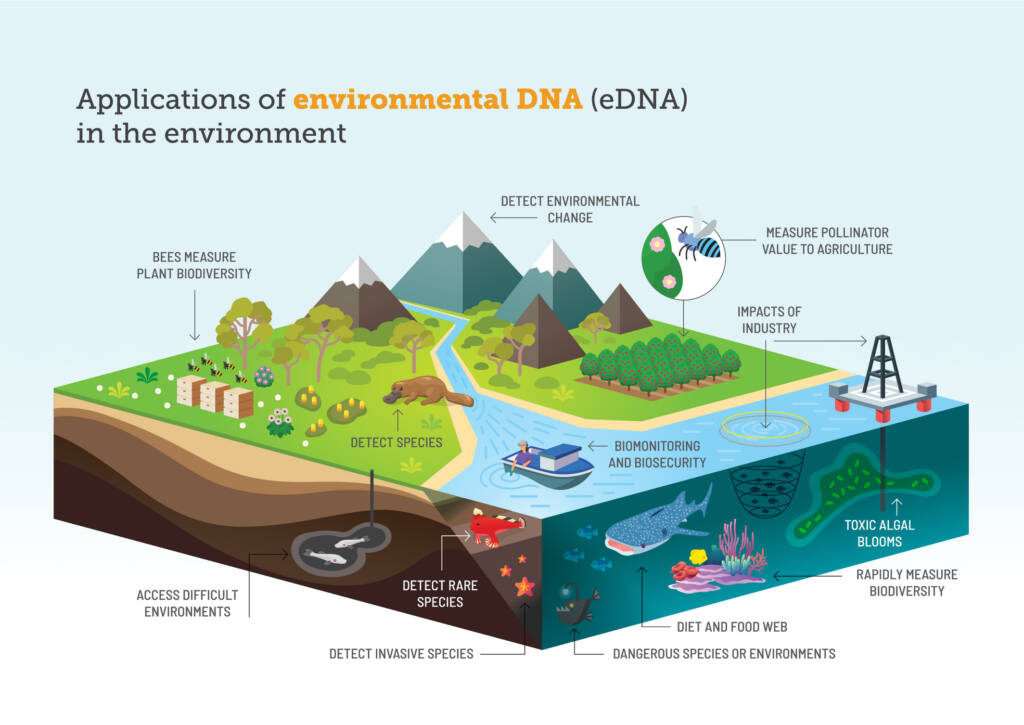
Applications of eDNA
eDNA is becoming more commonly used as a tool to complement baseline surveys and impact assessments. The data generated can be used to determine species presence, loss of species biodiversity, changes in species assemblages or compositions and introduction of invasive species. It can also be used to monitor for the return of indicator species to comment on rehabilitation success [8].
The taxonomic identification of eDNA from environmental samples can be used to aid a number of environmental disciplines including [9, 10]:
- Trophic and community ecology (understanding functional diversity and ecosystem dynamics);
- Biomonitoring (establish baseline data to inform policy making, assess restoration trajectories, conserve priority species)
- Population genetics (assessing variance or diversity within and between populations);
- Invasion biology (early species detection);
- Ecotoxicology (detection of anthropogenic contamination, microbial source tracking, identification of pathogens and parasites); and
- Ecology (determining species distributions across various spatial and temporal scales).
Considerations for Planning and Sample Collection
The collection of eDNA is a relatively simple process; however, to ensure that the methodology is suited to the assessment purpose, to limit the risk of cross-contamination, and to maximise detection probability, it is important to consult with the analysing laboratory prior to designing the sampling program to:
- Determine if the target species can be identified from eDNA e.g., reptiles are extremely hard to detect from eDNA;
- Determine what substrate will be sampled and how much of that substrate will be required;
- Evaluate the volume and number of sample containers required;
- Determine if there are any special requirements for sample preservation based on the required assay methods;
- Ensure the appropriate refrigeration equipment will be available to store samples between collection and dropping off to the lab i.e., fridge/esky; and
- Establish a standard sample naming convention to ensure samples are not mixed up.
As with traditional surveys, the sampling design also needs to consider the total number of samples and replicates required, the time of year sampling is carried out with regard to the target species [11].
Prior to sample collection, it is important to ensure appropriate protocols are in place to prevent contaminating samples with foreign DNA from the environment or from other samples. Standard practice can involve sterile gloves to be worn and replaced between collecting each sample, a new sterile container to be used to collect each sample, and a method for decontaminating field equipment between samples such as a bleach bucket and ethanol wipes.

eDNA Analysis
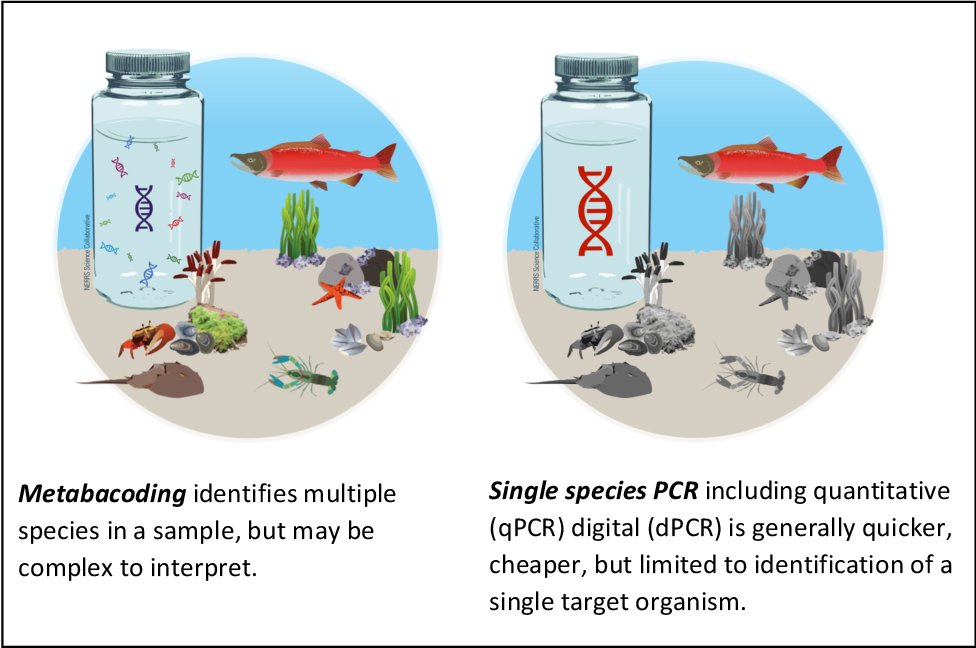
There are two different eDNA analytical options; single-species eDNA detection and multi-species eDNA detection. Single-species eDNA detection utilises species-specific markers to determine if a single species is present in the environmental sample [12]. This method is more reliable and accurate than multi-species detection and can be used to identify to species level. Multi-species eDNA detection, commonly known as metabarcoding, uses non-species-specific PCR primers to amplify a targeted region of DNA (barcode region) [5]. These amplicons are then sequenced and compared against a reference database for that barcode region to assign the amplicons to individual species [5].
eDNA Frontiers is the only laboratory in Western Australia that provides commercial eDNA services. The eDNA frontiers lab is located at Curtin University, and operates in conjunction with its sister lab, the Trace and Environmental DNA (TrEnD) laboratory, to enable industry, government and the public access to this innovative biodiversity tool.
Limitations and Industry Support
Despite the benefits associated with eDNA and substantial advancements the technology has undergone in the last two decades, there are a number of limitations that must be overcome before the method’s use prevails in conservation. Some of the limitations associated with the use of eDNA include:
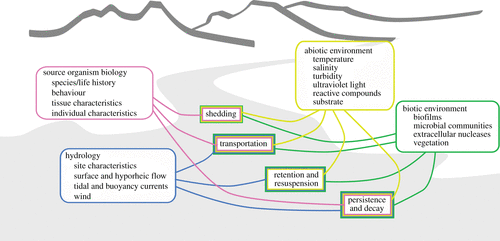
- Contamination – Contamination occurs when foreign DNA is contained in a sample that was taken from an environment where the individual that the DNA belongs to was not present at the time of sampling [4]. Contamination can occur in the field when sampling, e.g., eDNA from one location transferred to another location on clothing, and at various stages during sample processing in the lab, e.g., if a pipette tip is not replaced between handling different samples. In addition, in aquatic systems where eDNA methods are most commonly applied, foreign eDNA can be transported downstream or with currents, resulting in false positive detection of species in areas they are not actually present.
- DNA degradation – The persistence of eDNA varies significantly between different environments due to differences in abiotic/biotic conditions (microbial activity, UV, temperature and salinity), as well as between species, meaning eDNA may not be detected unless an individual has been in the sampled environment 1-2 weeks prior to sampling.
- Misidentification – To filter-out erroneous DNA, specific primers are used to target barcode regions of DNA that enable species identification from that targeted barcode sequence. DNA degradation or inconsistencies in the barcode region between species can result in misidentification. In addition, false negative detections can occur where no genetic material is present in the environmental sample, e.g., reptiles are notoriously difficult to detect due to limited deposition of genetic material [13].
- Species abundance – DNA shedding and degradation can be highly variable between species, which in turn can obscure the relationship between DNA concentration and species abundance. Stress, age, diet, temperature and community structure cause significant variation in volume and rate of tissue shedding [14]. eDNA can therefore not be used to provide accurate abundance information.
- Reference Library/Database – Identification of DNA sequences derived from environmental samples relies on accurate and complete reference databases of assigned DNA sequences. Where databases are complete and a high number of species are correctly detected in an environmental sample, eDNA methods can be extremely informative [15].However, where reference databases are incomplete, eDNA methods are far less effective as amplicons may be assigned to the wrong species, or to a lower biological classification level [16, 17].
Assigning sequences to species in reference databases is done using verified open-source data such as GenBank (https://www.ncbi.nlm.nih.gov/genbank/). To overcome the shortcomings of incomplete reference databases, there are a number of active projects around the world focused on increasing the amount of reference material available for use including the Earth Biogenome Project, Earth Holo Genome Initiative and the International Barcode of Life. Industry can assist by submitting or giving permission for the data they have collected to be added to genetic material reference systems [18]. By working together to improve the application of eDNA methods, this technology will become more effective and available for numerous applications across multiple industries.
Integrate Sustainability has a wealth of experience and knowledge in the management and coordination of baseline environmental surveys and environmental impact assessments. Integrate Sustainability actively looks for new technologies to provide our clients with appropriate, effective and cost-efficient solutions to aid, support or develop their projects. Should you need assistance with baseline survey management, impact assessments or are interested in finding out if eDNA analysis could be used on your project, please call us on (+618) 9468 0338, or contact us via email: enquiries@integratesustainability.com.au.
References
| [1] | A. V. Williams , P. G. Nevill and S. L. Krauss, “Next generation restoration genetics: applications and opportunities.,” Trends in Plant Sciences, vol. 19, no. 8, pp. 529-537, 2014. |
| [2] | B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, “The STructure and Function DNA,” in Molecular Biology of the Cell, 4 ed., New York, Garland Science, 2002. |
| [3] | D. S. Pilliod, C. S. Goldberg, M. B. Laramie and L. P. Waits, “Application of environmental DNA for inventory and monitoring of aquatic species: Fact Sheet 2012-3146,” U.S. Geological Survey, 2013. |
| [4] | E. M. Furlan and R. P. Duncan, “Identifying error and accurately interpreting environmental DNA metabarcoding results: A case study to detect vertebrates at arid zone waterholes.,” Molecular Ecology Resources, vol. 20, pp. 1259-1276, 2020. |
| [5] | P. Taberlet, E. Coissac, F. Pompanon, C. Brochmann and E. Willerslev, “Towards next-generation biodiversity assessment using DNA metabarcoding,” Molecular Ecology, vol. 21, pp. 2045-2050, 2012. |
| [6] | R. P. kelly, J. A. Port, K. M. Yamahara, R. G. Martone, N. Lowell, P. F. Thomsen, M. E. Mach, M. Bennet, E. Prahler, M. R. Caldwell and L. B. Crowder, “Harnessing DNA to improve environmental management: Genetic monitoring can help public agencies implement environmental laws,” Science, vol. 344, no. 6191, pp. 1455-1456, 2014. |
| [7] | M. A. Barnes and C. R. Turner, “The ecology of environmental DNA and implications for conservation genetics,” Conservation Genetics, vol. 17, pp. 1-17, 2016. |
| [8] | eDNA Frontiers, “Environmental Impact Assessment,” 2020. [Online]. Available: https://scieng.curtin.edu.au/edna-frontiers/environmental-impact-assessment/. [Accessed March 2021]. |
| [9] | D. Garlapati, B. Charankumar, K. Ramu, P. Madeswaran and M. V. Ramana Murthy, “A review on the applications and recent advances in environmental DNA (eDNA) metagenomics,” Reviews in Environmental Science and Bio/Technology, vol. 18, pp. 389 – 411, 2019. |
| [10] | E. E. Diaz-Ferguson and G. R. Moyer, “History, applications, methodological issues and perspectives for the use of environmental DNA (eDNA) in marine and freshwater environments,” Revista de Biolgia Tropical, vol. 62, no. 4, pp. 1273 – 1274, 2014. |
| [11] | E. F. McColl-Gausden, How to Guide: eDNA sampling for native, threatened and invasive species, Threatened Species Recovery Hub, National Environmental Science Programme, 2020. |
| [12] | S. Peixeto, C. Chaves, G. Velo-Antón, P. Beja and B. Egeter, “Species detection from aquatic eDNA: Assessing the importance of capture methods,” Environmental DNA, vol. 3, pp. 435-448, 2020. |
| [13] | G. F. Ficetola, R. Manenti and P. Taberlet, “Environmental DNA and metabarcoding for the study of amphibians and reptiles: species distribution, the microbiome, and much more,” Amphibia-Reptilia, vol. 40, no. 2, pp. 129-148, 2019. |
| [14] | J. B. Harrison, J. M. Sunday and S. M. Rogers, “Predicting the fate of eDNA in the study environment and impactions for studying biodiversity,” Proceedings of the Royal Society, vol. 286, no. 1915, 2019. |
| [15] | K. A. Thompson and S. G. Newmaster, “Molecular taxonomic tools provide more accurate estimates of species richness at less cost than traditional morphology-based taxonomic practices in a vegetation survey,” Biodiversity and Conservation, vol. 23, no. 6, pp. 1411-1424, 2014. |
| [16] | T. Bradford, M. Adams, W. F. Humphreys, A. D. Austin and S. J. B. Cooper, “DNA barcoding of stygofauna uncovers cryptic amphipod diversity in a calcrete aquifer in Western Australia’s arid zone,” Molecular Ecology Resources, vol. 10, pp. 41-50, 2010. |
| [17] | G. Carrasco-Puga, F. P. Diaz, D. C. Soto, C. Hernández-Castro, O. Conteras-López, A. Maldonado, C. Latorre and R. A. Gutierrez, “Revealing hidden plant diversity in arid environments,” Ecography, vol. 44, pp. 98-111, 2021. |
| [18] | O. Berry, S. Jarman, A. Bissett, M. Hope, C. Paeper, C. Bessey, M. K. Schwartz, J. Hale and M. Bunce, “MAking environmental DNA (eDNA) biodiversity records globally accessible,” Environmental DNA, vol. 3, pp. 699-705, 2021. |
| [19] | EcoDNA, “Technologies: Single Species Detection and Multi-Species Detection,” University of Canberra, 2019. |
| [20] | P. F. Thomsen and E. Willerslev, “Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity,” Biological Conservation, vol. 183, pp. 4-18, 2014. |
| [21] | C. Turner, “Environmental DNA in a Regulatory and Operational Landscape,” 2019. [Online]. Available: https://esassoc.com/news-item/environmental-dna-in-a-regulatory-and-operational-landscape/. |
| [22] | T. Simmons, D. Menning and S. Talbot, “Environmental DNA: An emerging tool for understanding aquatic biodiversity,” Alaska Park Science, vol. 19, no. 1, pp. 34-41, 2020. |
| [23] | O. Berry, S. Jarman, A. Bissett, M. Hope, C. Paeper, C. Bessey, M. K. Schwartz, J. Hale and M. Bunce, “Making environmental DNA (eDNA) biodiversity records globally accessible,” Environmental DNA, vol. 00, pp. 1-7, 2020. |
| [24] | A. Watts and J. Miksis-Olds, “The Ocean as a Living Sensor: Environmental DNA and Acoustics for Detecting Marine Life,” in The National Conference on Marine Environmental DNA, New York City, 2018. |
| [25] | Environmental Science Associates, “Environmental DNA in a Regulatory and Operational Landscape,” September 2019. [Online]. Available: https://esassoc.com/news-and-ideas/2019/09/environmental-dna-in-a-regulatory-and-operational-landscape/. |

