Throughout history, natural changes in the environment have contributed to the extinction of flora and fauna. However, disturbances created by humans is amplifying natural extinction rates reducing plant and animal populations worldwide. As a result, it is important that efforts are made to protect and conserve our planet’s biodiversity (Thomsen & Willerslev, 2015).
Biodiversity conservation is greatly dependant on successfully monitoring species distribution and populations size. Traditional monitoring techniques that surveying species richness and abundance rely on the physical capture and identification of species. These traditional methods can be hampered by taxonomic identification. They are invasive, time-consuming and prone to human errors. Some of these methods make it impossible to detect small and elusive species. Alternative techniques such as environmental DNA (eDNA), are increasingly being used in biodiversity surveys and for monitoring. eDNA is more efficient, non-invasive and cost-effective (Ruppert, et al., 2019).

What is Environmental DNA (eDNA)?
Environmental DNA (eDNA) is the process of extracting DNA samples from water, soil, snow or air, without distinctly identifying any particular organism (Seymour, 2019). Species can be classified and identified from the collected samples after they undergo the process of preservation, extraction, amplification, sequencing and categorisation.
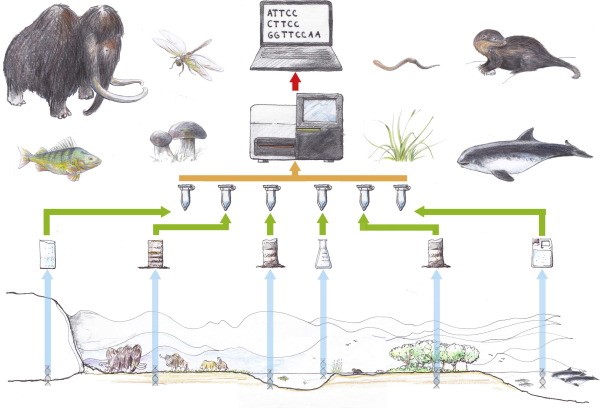
Samples of DNA can be extracted from numerous sources such as urine, carcasses, skin, blood, mucus, faeces, saliva, roots, fruit or pollen and in some instances microorganism are captured in entirety (Ruppert, et al., 2019). Excreted material can remain in the environment for a couple of days to a thousand years, depending on the preserving qualities of the environment. For example, samples in water would degrade quicker than those in sediments or permafrost (Whittington, 2015). The diagram below shows a workflow of the processing of samples and species which have been identified in their environment using eDNA (Thomsen & Willerslev, 2015).
eDNA uses, achievements and limitations
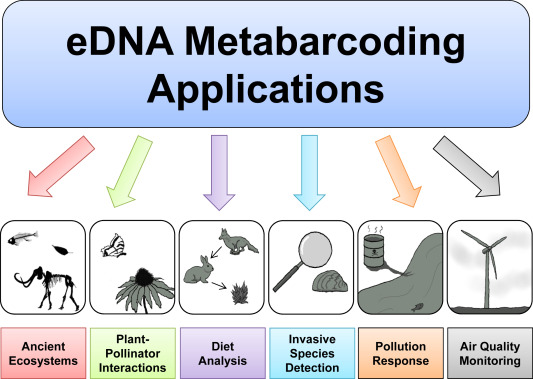
eDNA has been applied and used to answer questions not only in conservation biology but palaeontology and ecology as well. Species can be detected using either eDNA metabarcoding, where multiple species can be identified all at once, or by using quantitative PCR methods for single species detection (Thomsen & Willerslev, 2015). eDNA is a simpler sampling approach which can be carried out in most weather conditions (Whittington, 2015). It has proven to be the better method in species detection because it is a non-disruptive method; it does not cause any harm to the species or its habitat.
Elusive and rare species which can sometimes be impossible to identify physically in the field can be identified using eDNA. It also facilitates the early detection of invasive species. Additionally, eDNA enables researchers to observe species occurring in low densities, which are somewhat endangered or thought to be extinct (Ruppert, et al., 2019).
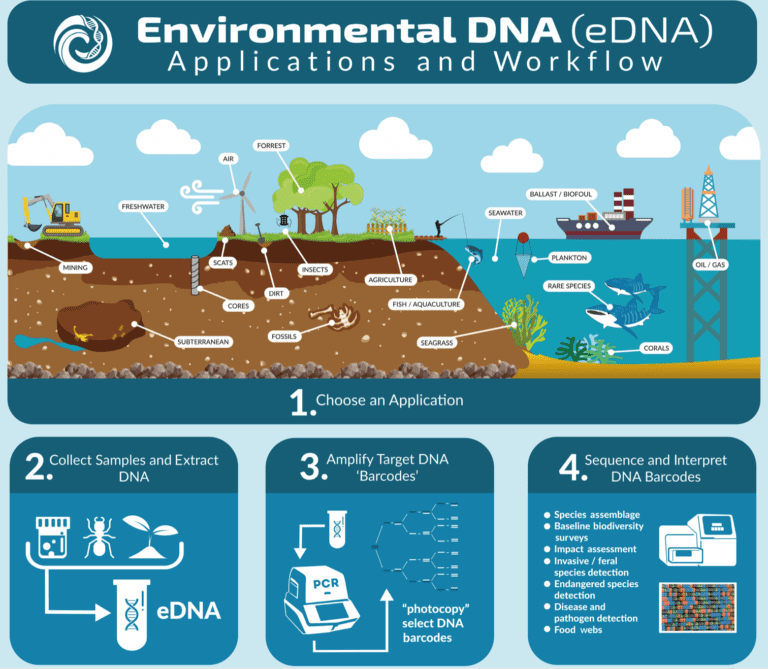
eDNA technology has opened many research opportunities globally. In Australia’s eastern rainforests, eDNA is being used to identify plants, animals, fungi, bacteria and archaea species from soil samples in the effort to explore biogeographic patterns (the distribution of organisms in space and time) (The Royal Botanic Garden Sydney, 2019). On the other side of the world, in Greenland, ice cores are being used to collect eDNA samples of ancient ecosystems (Hawley, 2018). New eDNA technology is assisting Hawaiian researchers monitor coral reef ecosystems. Coral Reefs provide habitat and critical resources to many other species but are unfortunately highly sensitive to the environmental changes caused by climate change. eDNA could potentially be a useful tool in preserving these unique coral ecosystems (University of Hawaii at Monoa, 2019).
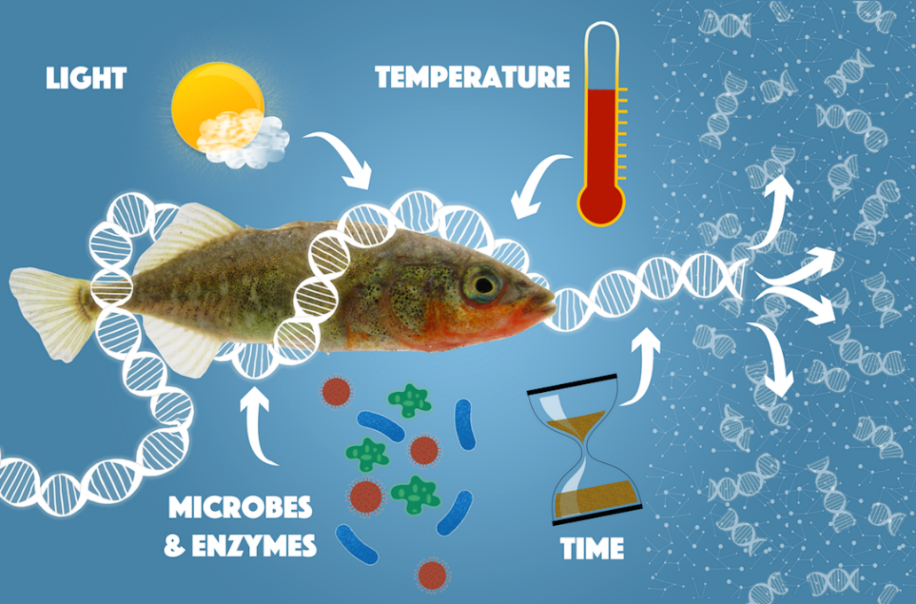
However, eDNA also has some shortfalls such as the risk of contamination, inhibition and sequencing errors which may produce false positives and inaccurate results (Thomsen & Willerslev, 2015). With eDNA, it is impossible to detect if the species identified was dead or alive at the time of excretion, or how long ago the species stayed in that environment. For example, eDNA can be preserved in soil for decades (or longer), so it makes it hard to identify if an organism was recently in the environment, or had been many years ago (Whittington, 2015).
eDNA degrades and distributes quickly, especially in water environments and when exposed to environmental factors such as light, heat, oxygen, microbes and enzymes. Through degradation, samples of DNA strands can become too short to provide a unique genetic fingerprint (Fishbio, 2015). Hence the collection of samples in some environments becomes time-sensitive.
eDNA is a very valuable tool for the detection and monitoring of species for conservation. However, at this stage eDNA can only be used as a complementary tool to the already existing traditional methods of species detection. Despite its limitations, eDNA possesses strengths which should be celebrated because it is a fast, efficient and non-invasive method that has provided researchers with invaluable information on many species, their behavioural patterns and how they interact with their environment.
Consider if eDNA is a tool that you can use to identify and monitor biodiversity on your project.
References
Biomeme, 2018. A Guide to
environmental DNA (eDNA). [Online]
Available at: https://biomeme.com/environmental-dna/
[Accessed 22 October 2019].
Fishbio, 2015. True Or False? Challenges Of EDNA Species
Detection. [Online]
Available at: https://fishbio.com/field-notes/the-fish-report/true-or-false-challenges-of-edna-species-detection
[Accessed 22 October 2019].
Genetics Home Reference, 2019. U.S. National Library of
Medicine. [Online]
Available at: https://ghr.nlm.nih.gov/primer/basics/dna
[Accessed 22 October 2019].
Ruppert, K. M., Kline, R. J. & Rahman, M. S., 2019. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Ecology and Conservation, Volume 17.
Seymour, M., 2019. Rapid progression and future of
environmental DNA research. [Online]
Available at: https://www.nature.com/articles/s42003-019-0330-9
[Accessed 22 October 2019].
The Guardian, 2019. DNA discussion at annual P.E.I.
genealogical meeting. [Online]
Available at: https://www.theguardian.pe.ca/lifestyles/local-lifestyles/dna-discussion-at-annual-pei-genealogical-meeting-356956/
[Accessed 22 October 2019].
Thomsen, P. F. & Willerslev, E., 2015. Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation, Volume 183, pp. 4-18.
Whittington, K., 2015. eDNA – the next big thing in
conservation monitoring?. [Online]
Available at: https://www.nature.com/scitable/blog/eyes-on-environment/edna_the_next_big_thing/
[Accessed 22 October 2019].



